Abstract
The C-terminal hypervariable region (HVR) of Ras proteins specifies post-translational modifications required for their subcellular localization and biologic function. The HVR of all four Ras isoforms terminates with a CAAX motif that is prenylated by farnesyltransferase, producing a weak membrane binding affinity that is further stabilized by a second signal motif. For K-Ras4b, this signal is provided by a polybasic lysine domain that allows direct localization of the protein at the plasma membrane (PM). By contrast, H-Ras, N-Ras, and K-Ras4a PM localization requires palmitoylation at cysteine(s) adjacent to the CAAX motif and is maintained through a dynamic cycle regulated by depalmitoylation and repalmitoylation. Importantly, these differences raise the provocative possibility that inhibiting palmitoylation could selectively target NRAS mutant cancer cells without toxicity to normal cells that retain intact K-Ras4b function.
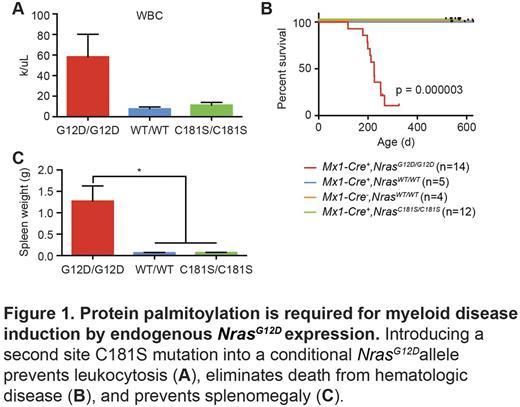
Indeed, Nras oncogenes with a cysteine-to-serine second site amino acid substitution at the palmitoylation site (C181S) exhibited defective transforming activity in hematopoietic stem and progenitor cells (HSPCs) ex vivo and in vivo (Cuiffo et al., Blood 2010; Xu et al., Blood 2012). However, these studies relied on retroviral promoters to express oncogenic N-RasG12D that resulted in supra-physiological protein levels in HSPCs. To overcome this limitation, we retargeted embryonic stem cells that were used to generate Lox-STOP-Lox (LSL) NrasG12D/+ knock in mice ( Haigis et al., Nat Genet 2008 ) to create a NrasG12D,C181S allele (hereafter called NrasC181S). We then performed intercrosses to generate Mx1-Cre, NrasC181S/C181S mice as well as cohorts of congenic Mx1-Cre, NrasWT/WT and Mx1-Cre, NrasG12D/G12D control animals. As expected (Li et al ., Blood 2011; Wang et al ., Cell Cycle 2011), Mx1-Cre, NrasG12D/G12D mice that were injected with poly(I:C) at weaning had a mean white blood cell (WBC) count of 58.3 x 103/µL at six months of age (Fig.1 A) and died from hematologic disease after 225 days (range 119-267 days, n= 14, Fig. 1B). By contrast, Mx1-Cre, NrasC181S/C181S mice had normal WBC counts at the same time point (mean: 11.34x103 WBC/µL) and remained well until 19-20 months of age (n=12). The difference in survival between these groups was highly significant (p=3x10-6 by Kaplan-Meier analysis). Detailed analysis of the hematopoietic compartment at six months of age (n=5 per genotype) revealed profound splenomegaly (mean weight: 1.272g) in NrasG12D/G12D mice, but normal spleen weights in the NrasWT/WT and NrasC181S/C181S animals (Fig. 1C). The expansion of granulocyte-monocyte progenitors (GMPs), megakaryocyte-erythroid progenitors (MEPs), and more primitive CD48+ Lin- c-Kit+ Sca1+ (LKS) cells previously observed in NrasG12D/G12D mice was also rescued by the C181S second site mutation. Furthermore, in vitro culture of HSPC-enriched bone marrow cells over a range of granulocyte-macrophage colony-stimulating factor (GM-CSF) concentrations revealed normal cytokine sensitivity of NrasC181S/C181S cells, while NrasG12D/G12D cells were characterized by cytokine-independent growth and GM-CSF hypersensitivity as previously described. Importantly, Ras-GTP levels were similarly elevated in bone marrow cells from both NrasG12D/G12D and NrasC181S/C181S mice compared to wild-type cells. This indicates that the C181S mutation abrogates the cytokine hypersensitivity and increased cell proliferation that results from oncogenic N-RasG12D expression without reverting the canonical biochemical consequence of the oncogenic G12D substitution. This observation, in turn, suggests that abnormal subcellular localization and trafficking of oncogenic N-Ras potently suppresses myeloid transformation. Taken together, these in vivo studies directly validate the palmitoylation/depalmitoylation cycle as a therapeutic target in hematologic malignancies and potentially in other cancers characterized by oncogenic NRAS mutations.
Firestone: Calico Life Sciences: Employment.
Author notes
Asterisk with author names denotes non-ASH members.